Plot choropleth map.
Usage
mf_choro(
x,
var,
pal = "Mint",
alpha = 1,
rev = FALSE,
breaks = "quantile",
nbreaks,
border = getOption("mapsf.fg"),
pch = 21,
cex = 1,
lwd = 0.7,
col_na = "white",
cex_na = 1,
pch_na = 4,
leg_pos = mf_get_leg_pos(x),
leg_title = var,
leg_title_cex = 0.8,
leg_val_cex = 0.6,
leg_val_rnd = 2,
leg_no_data = "No data",
leg_frame = FALSE,
leg_horiz = FALSE,
leg_adj = c(0, 0),
leg_size = 1,
leg_box_border = getOption("mapsf.fg"),
leg_box_cex = c(1, 1),
leg_fg = getOption("mapsf.fg"),
leg_bg = getOption("mapsf.bg"),
leg_frame_border = getOption("mapsf.fg"),
add = FALSE
)Arguments
- x
object of class
sf- var
name(s) of the variable(s) to plot
- pal
a set of colors or a palette name (from hcl.colors)
- alpha
if
palis a hcl.colors palette name, the alpha-transparency level in the range [0,1]- rev
if
palis a hcl.colors palette name, whether the ordering of the colors should be reversed (TRUE) or not (FALSE)- breaks
either a numeric vector with the actual breaks, or a classification method name (see mf_get_breaks and Details)
- nbreaks
number of classes
- border
border color
- pch
pch type of pch if x is a POINT layer
- cex
cex cex of the symbols if x is a POINT layer
- lwd
border width
- col_na
color for missing values
- cex_na
cex for NA values if x is a POINT layer
- pch_na
pch for NA values if x is a POINT layer
- leg_pos
position of the legend, one of 'topleft', 'top','topright', 'right', 'bottomright', 'bottom', 'bottomleft', 'left' or a vector of two coordinates in map units (c(x, y)). If leg_pos = NA then the legend is not plotted. If leg_pos = 'interactive' click onthe map to choose the legend position.
- leg_title
legend title
- leg_title_cex
size of the legend title
- leg_val_cex
size of the values in the legend
- leg_val_rnd
number of decimal places of the values in the legend
- leg_no_data
label for missing values
- leg_frame
whether to add a frame to the legend (TRUE) or not (FALSE)
- leg_horiz
display the legend horizontally (for proportional symbols and choropleth types)
- leg_adj
adjust the postion of the legend in x and y directions
- leg_size
size of the legend; 2 means two times bigger
- leg_box_border
border color of legend boxes
- leg_box_cex
width and height size expansion of boxes
- leg_fg
color of the legend foreground
- leg_bg
color of the legend backgournd
- leg_frame_border
border color of the legend frame
- add
whether to add the layer to an existing plot (TRUE) or not (FALSE)
Details
Breaks defined by a numeric vector or a classification method are
left-closed: breaks defined by c(2, 5, 10, 15, 20)
will be mapped as [2 - 5[, [5 - 10[, [10 - 15[, [15 - 20].
The "jenks" method is an exception and has to be right-closed.
Jenks breaks computed as c(2, 5, 10, 15, 20)
will be mapped as [2 - 5], ]5 - 10], ]10 - 15], ]15 - 20].
Examples
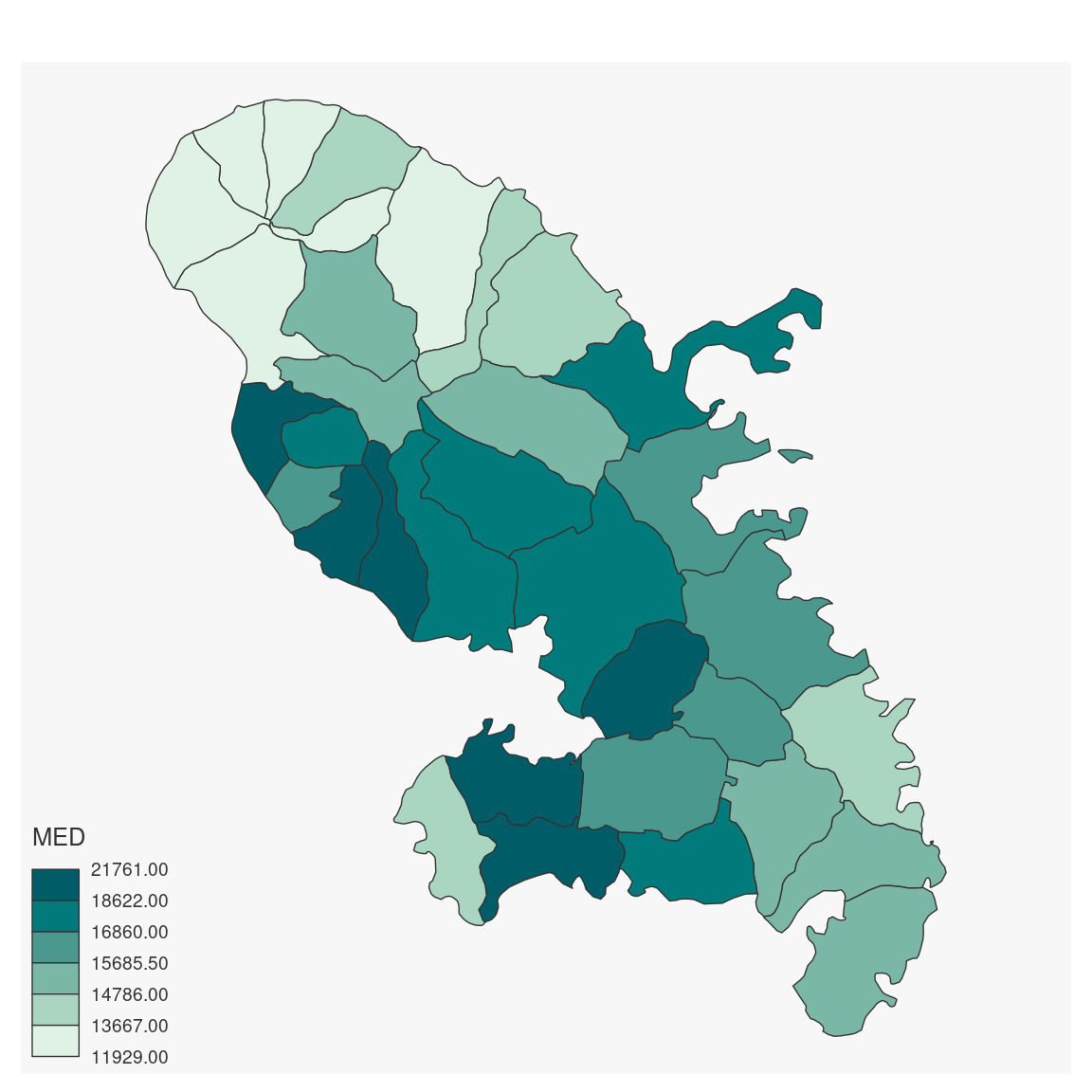
mtq <- mf_get_mtq()
mf_map(mtq, var = "MED", type = "choro")
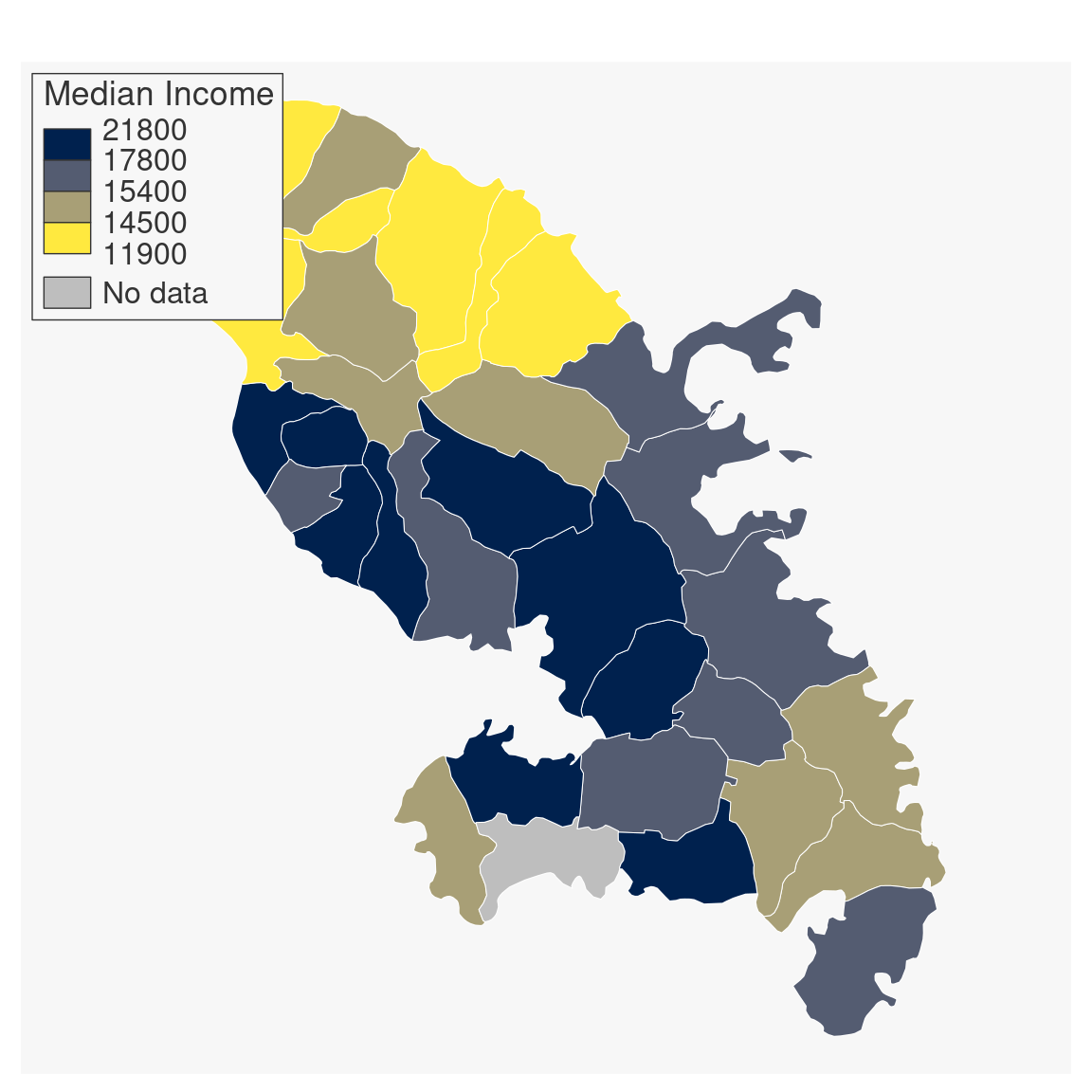
 mtq[6, "MED"] <- NA
mf_map(
x = mtq, var = "MED", type = "choro",
col_na = "grey", pal = "Cividis",
breaks = "quantile", nbreaks = 4, border = "white",
lwd = .5, leg_pos = "topleft",
leg_title = "Median Income", leg_title_cex = 1.1,
leg_val_cex = 1, leg_val_rnd = -2, leg_no_data = "No data",
leg_frame = TRUE
)
mtq[6, "MED"] <- NA
mf_map(
x = mtq, var = "MED", type = "choro",
col_na = "grey", pal = "Cividis",
breaks = "quantile", nbreaks = 4, border = "white",
lwd = .5, leg_pos = "topleft",
leg_title = "Median Income", leg_title_cex = 1.1,
leg_val_cex = 1, leg_val_rnd = -2, leg_no_data = "No data",
leg_frame = TRUE
)